This Bioinformatics Protocols page contains a in silico protocols intended for use in higher education teaching and academic research. The protocols were developed by Prof Nat Milton in his academic and commercial roles.
The attached Summary Protocol and Full Protocol have specifically been written by Prof Milton, who has used antisense peptide techniques for both academic research and commercial activities with the his academic research group and with NeuroDelta Ltd. The methods detailed in the protocol are aimed at University students plus academic staff for use in non-commercial activities. The main method is based around antisense peptide screening [1] and has been adapted to protein sequence database screening [2]. Support for these Bioinformatics protocols from is available directly via Email.
An example of the use of these techniques was a study to identify Alzheimer's amyloid-ß (Aß) binding peptides that could block both neurotoxicity and fibril formation of Aß. The binding peptides were developed based on generating antisense peptides that would specifically bind the Aß 1-43 peptide using the messenger RNA (mRNA) sequence coding for the peptide as a template [3],[4]. The Aß antisense peptide sequences and data from their generation plus characterisation are the subject of published patent applications [4] and were used to set up the NeuroDelta Ltd company for commercial exploitation of these patents. Subsequent studies by other groups demonstrated that similar Aß antisense peptides were able to prevent the toxicity of Aß [5], confirming the original observations.
The use of the Aß 1-43 antisense peptide screening lead to the identification of a catalase-Aß binding domain, plus development of compound R9 that was used to block the catalase-Aß interaction [6],[7]. The use of the Aß 1-43 antisense peptide screening also lead to the identification of the Kissorphin (KSO) peptide [8] as an amyloid binding compound that was effective against Aß, amylin and prion protein toxicity [9],[10].
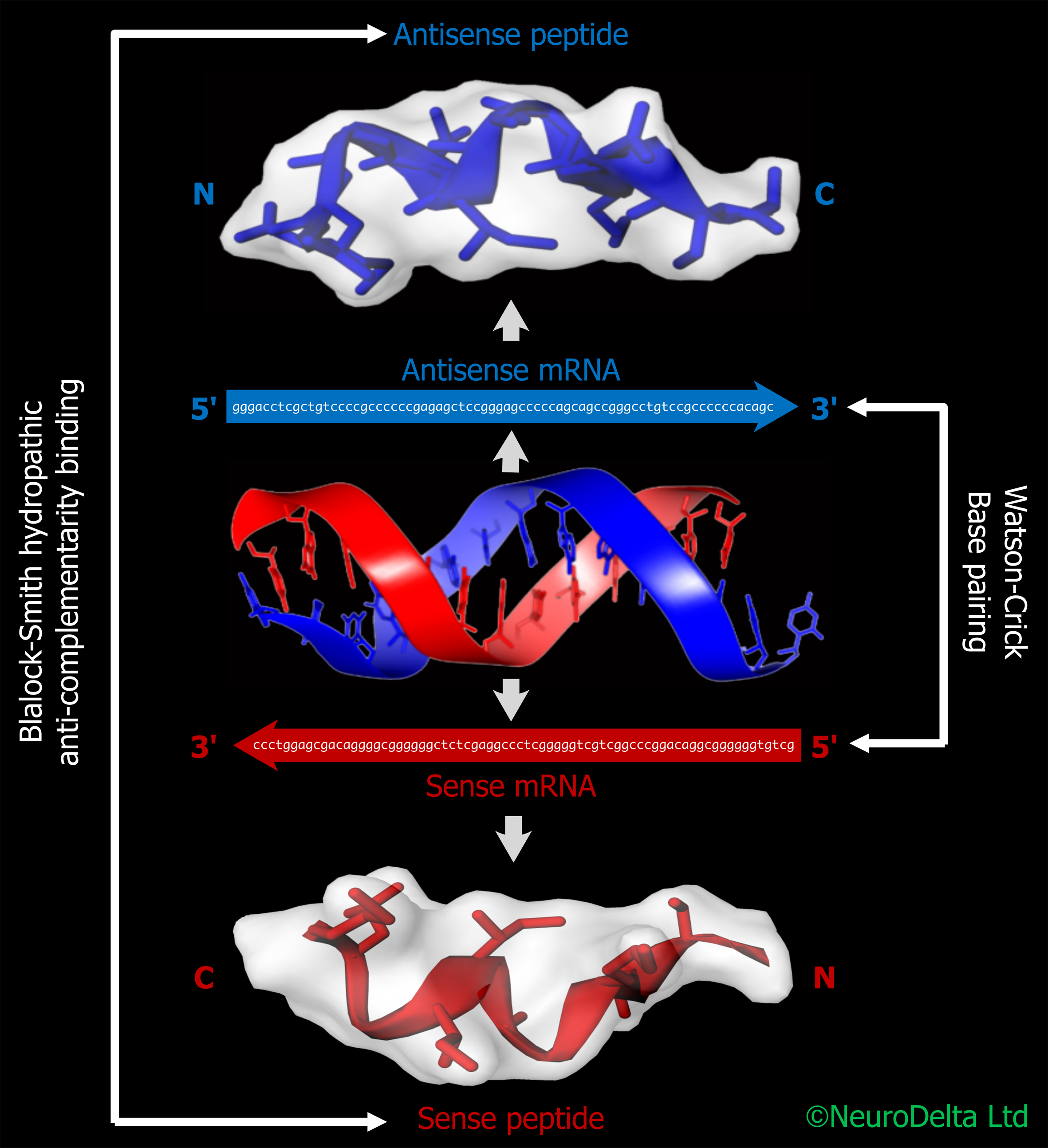
The protocols detail how to use an online method (using a Python script or Word version which can be used in an online Python Compiler or a downloaded version of Python 3) to generate antisense peptide sequences from a target protein mRNA sequence and has been based on the methods outlined in [2],[11],[12]. Methods using the antisense peptides in protein BLAST searches to identify potential protein interactions with the target protein are included. Protein-protein docking of 3D models can be used to predict the structure of complexes between the proteins identified from the antisense peptide screening [13],[14].
The following links may be used to access software in conjunction with the protocol:
Bacterial and Viral Bioinformatics Resource Center
BLAST
ClusPro
EzMol - Molecular display wizard
GROMACS - Gromacs
iCn3D
I-TASSER server for protein structure and function prediction
ModelArchive
NCBI
PDB-101: Guide to Understanding PDB Data
PDBx/mmCIF conversion service
PubMed
Python compiler
Python script
RCSB PDB
RENAME CHAIN IN PDB FILE
The Human Protein Atlas
UniProt
Upside Down Text | Flip Text, Type Upside Down, or Backwards Text
ZDOCK